While Human Leukocyte Antigen (HLA) matching is often emphasized in the context of bone marrow and organ transplant, HLA typing is used to identify disease-related biomarkers and predict immune responses, including those that could lead to cell rejection and/or graft-versus-host disease (GvHD), which is why researchers are interested in HLA phenotyping and genotyping of cellular starting material. The major histocompatibility complex (MHC) region on chromosome 6, where the HLA genes are encoded, is one of the most complex and polymorphic regions in the human genome. Although there are more than 200 HLA genes, 6 are crucial for the distinction between self and non-self. HLA-A, HLA-B, and HLA-C genes/loci belong to class I, and HLA-DP, HLA-DQ, and HLA-DR belong to class II. More details about HLA basics can be found in our previous blog post.
HLA typing can be performed based on serology at the antigen level, which detects anti-HLA antibodies, or at the DNA level by sequencing, which provides specific allele information. Serology testing used to be the “gold standard” HLA typing, relying on histocompatibility assays which identify molecules that are actually expressed on the cells and are detected by monoclonal antibodies. It is quick, simple, and low cost test that established an HLA phenotype. However, serological HLA typing doesn’t account for sequence variation like some differences in the HLA-DR locus, which is important in organ and cord blood transplantation.
In order to identify polymorphism (DNA sequence/coding variation) at the HLA-DR gene for example, typing must be done at the genetic level. At this DNA level, specific sequencing is performed which distinguishes HLA type at the allelic resolution (alleles are the variant forms of a single gene). There are currently >30,000 known HLA alleles, each designated by a unique number comprised of at least four digits. The HLA allele number usually corresponds to the allele group and the HLA protein, while longer numbers are only assigned when necessary (Figure 1).
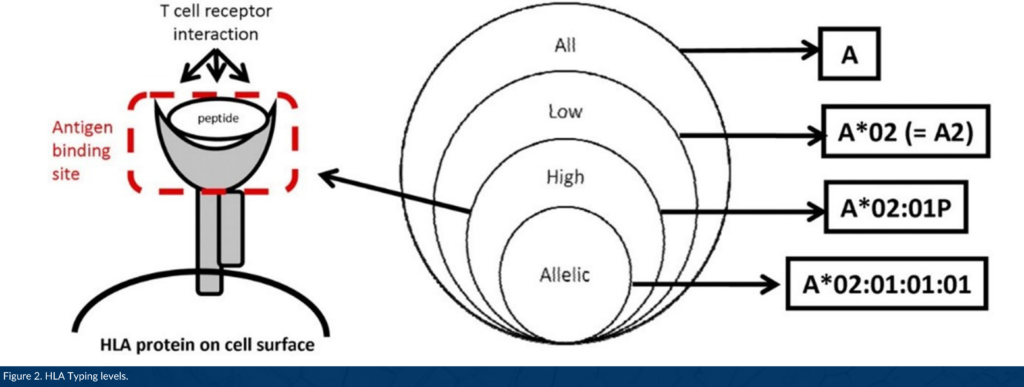
DNA-based HLA typing is often designated as low, intermediate, or high resolution. Low resolution typing corresponds to an allele group of a particular HLA gene (for example, A*01; A*02). High resolution typing corresponds to a set of alleles that encode one specific HLA protein region called the antigen binding site, used to present a peptide to immune cells (for example, A*01:02; A*02:01, Figure 2).1 High resolution typing resolves ambiguities resulting from substitutions located within exons 2 and 3 for class I HLA genes, and exon 2 for class II genes located within coding regions. This level of high resolution is usually referred to as 1x resolution. When high resolution HLA typing includes non-coding regions and allele sequences that are not expressed as cell-surface proteins, high resolution typing is termed 2x, 3x, 4x, etc.
Along with other newly discovered biomarkers, HLA phenotype and genotype may be used to predict therapeutic outcomes in a wide range of indications, including cancer. In a recent study analyzing thousands of immuno-oncology patients, researchers from the National Cancer Institute found a link between HLA-A*03 allele carriers and poor response to different checkpoint inhibitor treatments in multiple types of cancer.2 This is one of many examples of how top-quality, highly characterized cellular starting materials enable researchers with accurate donor selection and help clients move efficiently through the pre-clinical, clinical, and commercial phases of development to advance the delivery of life-saving, breakthrough therapies.
All of OrganaBio’s cell products are HLA typed by next (third) generation sequencing (NGS) of all 6 HLA genes and high resolution results are reported. High resolution genotyping by NGS is performed for HLA-A, B, C, DR, DQ, DP, and scientists can pick cell products from donors with common or rare HLA alleles. Request a copy of our inventory report here.

A description of the parts in an HLA name. Allele group; the allele’s antigen type, typically found by serotyping. HLA protein; the peptide for which the allele codes. Synonymous DNA substitution in a coding region; an allele variant with a different DNA sequence that produces the same protein. DNA substitution in a non-coding region; identifies a sequence polymorphism/mutation outside of the coding region.
References
- Nunes E, Heslop H, Fernandez-Vina M, Taves C, Wagenknecht DR, Eisenbrey AB, Fischer G, Poulton K, Wacker K, Hurley CK, Noreen H, Sacchi N. Definitions of histocompatibility typing terms. Blood. 2011 Dec 1;118(23):e180-3.
- Naranbhai V, Viard M, Dean M, Groha S, Braun DA, Labaki C, Shukla SA, Yuki Y, Shah P, Chin K, Wind-Rotolo M, Mu XJ, Robbins PB, Gusev A, Choueiri TK, Gulley JL, Carrington M. HLA-A*03 and response to immune checkpoint blockade in cancer: an epidemiological biomarker study. Lancet Oncol. 2022 Jan;23(1):172-184.






